Old Browser
Your account has been put on hold due to inactivity. To re-activate, check your account information and make all necessary updates.
Looks like you're visiting us from {{countryName}}.
Would you like to remain on the current country site or be redirected to one based on your location?
Your account has been put on hold due to inactivity. To re-activate, check your account information and make all necessary updates.
Overview
Analyzing paired chain information of T cell receptors (TCR) and B cell receptors (BCR) at the single-cell level is a powerful tool for probing T and B cell diversity and function in lymphoid malignancies, infectious diseases, autoimmune disorders and tumor immunology. The BD Biosciences TCR and BCR profiling tools help you uncover clonal diversity and function of T and B cells, respectively, at the single-cell level.
BD Rhapsody™ TCR/BCR Multiomic Assay for Human and Mouse
The BD Rhapsody™ TCR/BCR Multiomic Assay helps you assemble the full length VDJ sequence for the T cell and B cell receptor while assessing transcriptomic and protein expression of the same cells. The assay can serve as the one-stop shop for your single-cell multiomics needs.
The BD Rhapsody™ TCR/BCR Multiomic Assay for Human and Mouse includes:
- Full multiomics optimization and validation
- Reagents and pipeline to support assembly of full-length TCR and BCR sequences
- Compatibility with whole transcriptome and targeted mRNA assays
- Enhanced cell capture beads for sensitive molecule detection
BD Rhapsody™ VDJ CDR3 Protocol with V1 beads
The BD Rhapsody™ VDJ CDR3 Protocol utilizes the existing BD Rhapsody™ Targeted Kits and the human/mouse immune response primer panel and is designed to work alongside the BD® AbSeq Assay and BD® Single-Cell Multiplexing Kit (SMK). Details about the individual reagents needed to run the assay are included in the respective protocols. A dedicated bioinformatics pipeline is also available for you to analyze sequencing data generated using the CDR3 protocol.
The BD Rhapsody™ VDJ CDR3 Assay for profiling TCR and BCR includes:
- Validated primer sequences
- A detailed step-by-step protocol for analyzing the human or mouse CDR3 region of TCR and BCR using the BD Rhapsody™ Single-Cell Analysis System
Features
The BD Rhapsody™ TCR/BCR Multiomic Assay has been fully validated with our new enhanced cell capture beads as well as BD® AbSeq Antibody-Oligos and the BD® Single-Cell Multiplexing Kit (SMK). Confidently analyze the transcriptome, proteome and full length TCR/BCR repertoire of your cells, including gamma/delta T cells, from the only single-source provider of single-cell multiomics products.
Applications
A single workflow to profile the T cell and B cell receptors validated with existing targeted or WTA mRNA assays as well as protein analysis and sample multiplexing
In the experiment outlined below, resting human PBMCs, stained with the BD® AbSeq Immune Discovery Panel and four drop-in BD® AbSeq Antibodies, were multiplexed with enriched T cells stained with a 15-plex AbSeq panel. The pooled samples were processed with the TCR/BCR multiomic assay and WTA assay (Cat. No. 633801). The following example analysis was conducted on the T cells within the PBMC data.
Step 1:
Using both protein and gene expression a user can generate a tSNE plot and use AbSeq to identify cell populations.
tSNE plot (right) with major cell lineages identified. CD8 positive cells are highlighted and analyzed deeper as shown in subsequent figures.
Step 2:
An unsupervised analysis (phenograph) was performed on CD8 + cells where six clusters were identified
Six distinct CD8 positive subpopulations (right) were identified through an unsupervised phenograph analysis.
Step 3:
Frequencies of unique clonotypes within clusters were reported
Expanded paired TCR clonotypes for alpha and beta chains (right) were identified within phenograph 2 of CD8 positive cells.
Step 4:
The highest frequency clonotype was overlaid on the tSNE plot and identified within cluster two
The location of individual cells harboring the expanded paired clonotypes were identified by overlaying data on the tSNE plot (right).
Step 5:
Using AbSeq and WTA information, differential gene and protein expression analysis of cells with the most frequent clonotype vs the rest of the population can be performed
Gene and protein expression data for cells with the most frequent clonotype are compared to remaining CD8 positive cells (right).
Step 6:
Cell IDs and chain information can be obtained from pipeline output files
The cell label and CDR3 information for cells expressing the expanded clonotype were identified and used for further analysis.
| Cell_Index | Chain_Type | CDR3_Translation_Dominant |
|---|---|---|
| 54044 | TCR_Alpha | AVLAEGGKLI |
| 54044 | TCR_Beta | ASSLWGSPLH |
| 123296 | TCR_Alpha | AVLAEGGKLI |
| 123296 | TCR_Beta | ASSLWGSPLH |
| 123563 | TCR_Alpha | AVLAEGGKLI |
| 123563 | TCR_Beta | ASSLWGSPLH |
| 147961 | TCR_Alpha | AVLAEGGKLI |
| 147961 | TCR_Beta | ASSLWGSPLH |
Step 7:
VDJ and C gene information can be determined for clonotype of interest
The translated V(D)J gene segments for cells' labels of interest were tabulated from bioinformatic ouptut files.
| Cell_Index | Chain_Type | V_gene_Dominant | D_gene_Dominant | J_gene_Dominant | C_gene_Dominant | Full_Length | Productive |
|---|---|---|---|---|---|---|---|
| 54044 | TCR_Alpha | TRAV20*01 | TRAJ23*01 | TRAC | TRUE | TRUE | |
| 54044 | TCR_Beta | TRAV27*01 | TRBD1*01 | TRBJ1-6*02 | TRBC1 | TRUE | TRUE |
| 123296 | TCR_Alpha | TRAV20*01 | TRAJ23*01 | TRAC | TRUE | TRUE | |
| 123296 | TCR_Beta | TRAV27*01 | TRBD1*01 | TRBJ1-6*02 | TRBC1 | TRUE | TRUE |
| 123563 | TCR_Alpha | TRAV20*01 | TRAJ23*01 | TRAC | TRUE | TRUE | |
| 123563 | TCR_Beta | TRAV27*01 | TRBD1*01 | TRBJ1-6*02 | TRBC1 | TRUE | TRUE |
| 147961 | TCR_Alpha | TRAV20*01 | TRAJ23*01 | TRAC | TRUE | TRUE | |
| 147961 | TCR_Beta | TRAV27*01 | TRBD1*01 | TRBJ1-6*02 | TRBC1 | TRUE | TRUE |
Step 8:
Full length sequence of clonotype can be obtained (nucleotide sequence also available)
| Cell_Index | VDG_Translation_Trimmed |
|---|---|
| 54044 | EDQVTQSPEALRLQEGESSSLNCSYTVSGLRGLFWYRQDPGKGPEFLFTLYSAGEEKEKERLKATLTKKESFLHITAPKPEDSATYLCAVLAEGGKLIFGQGTELSVKP |
| 54044 | EAQVTQNPRYLITVTGKKLTVTCSQNMNHEYMSWYRQDPGLGLRQIYYSMNVEVTDKGDVPEGYKVSRKEKRNFPLILESPSPNQTSLYFCASSLWGSPLHFGNGTRLTVT |
| 123296 | EDQVTQSPEALRLQEGESSSLNCSYTVSGLRGLFWYRQDPGKGPEFLFTLYSAGEEKEKERLKATLTKKESFLHITAPKPEDSATYLCAVLAEGGKLIFGQGTELSVKP |
| 123296 | EAQVTQNPRYLITVTGKKLTVTCSQNMNHEYMSWYRQDPGLGLRQIYYSMNVEVTDKGDVPEGYKVSRKEKRNFPLILESPSPNQTSLYFCASSLWGSPLHFGNGTRLTVT |
| 123563 | EDQVTQSPEALRLQEGESSSLNCSYTVSGLRGLFWYRQDPGKGPEFLFTLYSAGEEKEKERLKATLTKKESFLHITAPKPEDSATYLCAVLAEGGKLIFGQGTELSVKP |
| 123563 | EAQVTQNPRYLITVTGKKLTVTCSQNMNHEYMSWYRQDPGLGLRQIYYSMNVEVTDKGDVPEGYKVSRKEKRNFPLILESPSPNQTSLYFCASSLWGSPLHFGNGTRLTVT |
| 147961 | EDQVTQSPEALRLQEGESSSLNCSYTVSGLRGLFWYRQDPGKGPEFLFTLYSAGEEKEKERLKATLTKKESFLHITAPKPEDSATYLCAVLAEGGKLIFGQGTELSVKP |
| 147961 | EAQVTQNPRYLITVTGKKLTVTCSQNMNHEYMSWYRQDPGLGLRQIYYSMNVEVTDKGDVPEGYKVSRKEKRNFPLILESPSPNQTSLYFCASSLWGSPLHFGNGTRLTVT |
The full-length amino acid sequence of the V(D)J alpha and beta TCR chains shown here were annotated showing framework and CDR regions in different colors.
| TCR_alpha | EDQVTQSPEALRLQEGESSSLNCSYTVSGLRGLFWYRQDPGKGPEFLFTLYSAGEE KEKERLKATLTKKESFLHITAPKPEDSATYLCAVLAEGGKLIFGQGTELSVKP |
| TCR_beta | EAQVTQNPRYLITVTGKKLTVTCSQNMNHEYMSWYRQDPGLGLRQIYYSMNVEVTDKGDVPEGYKVSRKEKRNFPLILESPSPNQTSLYFCASSLWGSPLHFGNGTRLTVT |
-
Protocols
-
BD Rhapsody™ System TCR/BCR Full Length and Targeted mRNA Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length, Targeted mRNA, and Sample Tag Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length, Targeted mRNA, and BD® AbSeq Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length, Targeted mRNA, BD® AbSeq, and Sample Tag Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length and mRNA Whole Transcriptome Analysis (WTA) Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length, mRNA Whole Transcriptome Analysis (WTA), and Sample Tag Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length, mRNA Whole Transcriptome Analysis (WTA), and BD® AbSeq Library Preparation
-
BD Rhapsody™ System TCR/BCR Full Length, mRNA Whole Transcriptome Analysis (WTA), BD® AbSeq, and Sample Tag Library Preparation
-
Brochure
FEATURES
The BD Rhapsody™ VDJ CDR3 Assay can be used with the BD Rhapsody™ Targeted Kit, BD® AbSeq Assay and BD® Single-Cell Multiplexing Kit, providing flexibility in your research
Based on the research need, the assay can be used in any of the following combinations:
- BD Rhapsody™ VDJ CDR3 Protocol—utilizes the BD Rhapsody™ Targeted Kit + Human Immune Response Panel* and enables you to analyze TCR and BCR sequences.
- BD Rhapsody™ VDJ CDR3 + SMK Protocol—utilizes the BD Rhapsody™ Targeted Kit + Human Immune Response Panel* + SMK and enables you to process multiple samples together in a single cartridge.
- BD Rhapsody™ VDJ CDR3 + BD® AbSeq Protocol—utilizes the BD Rhapsody™ Targeted Kit + Human Immune Response Panel* + BD® AbSeq Assay and enables multiomic analyses integrating TCR and BCR sequence analyses alongside protein information on the same sample.
- BD Rhapsody™ VDJ CDR3 + BD® AbSeq + SMK Protocol—requires the BD Rhapsody™ Targeted Kit + Human Immune Response Panel* + BD® AbSeq Assay + SMK and enables you to perform multiomic analyses on multiple samples of interest together in a single cartridge.
*While the VDJ protocol can technically work with any targeted panel, it works best with the immune response panel. To work effectively with our CDR3 VDJ assay, we recommend including immune response panel makers in custom panels.
PERFORMANCE
A powerful assay for analyzing CDR3 information of TCR and BCR
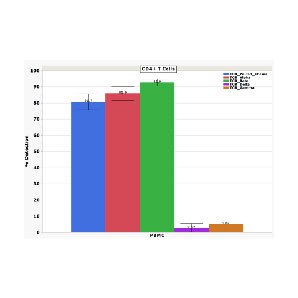
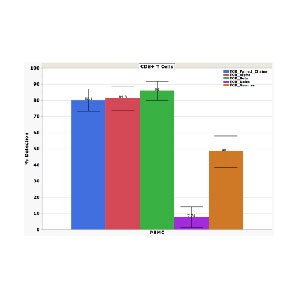
The hallmark of the BD Rhapsody™ VDJ CDR3 Protocol is its high sensitivity and specificity in detecting distinct clonotypes of T and B cells including gamma delta T cells alongside protein analyses.
High sensitivity and specificity in TCR detection
High paired TCR sensitivity in CD4+ T cells
High paired TCR sensitivity in CD8+ T cells

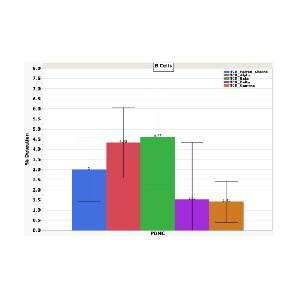
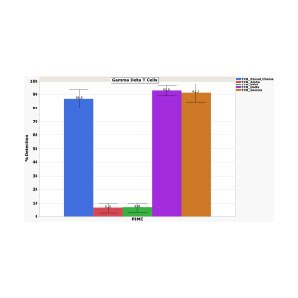
Detection of BCR, gamma delta T cells and protein markers
High sensitivity in identifying paired heavy and light chain information for BCR in B cells

Detection of gamma delta T cells to identify the rare and precious gamma delta T cells in your T cell population
APPLICATIONS
A multiomic tool for performing in-depth immune cell profiling
The BD Rhapsody™ VDJ CDR3 Protocol can be combined with the BD® AbSeq Assay and BD® Single-Cell Multiplexing Kit to perform multiomic analyses of both protein and RNA information. Here, the BD Rhapsody™ VDJ CDR3 Protocol was used to analyze the profile and clonotype amplification of T cells in response viral peptides.
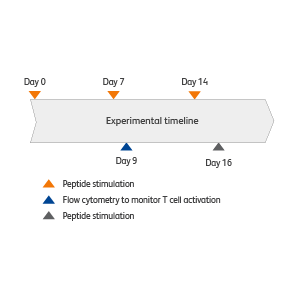

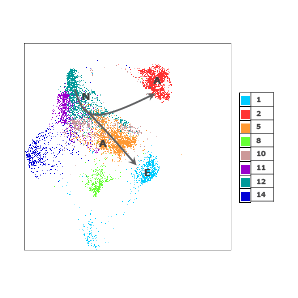
For Research Use Only. Not for use in diagnostic or therapeutic procedures.
Report a Site Issue
This form is intended to help us improve our website experience. For other support, please visit our Contact Us page.
